diff --git a/.github/CONTRIBUTING.md b/.github/CONTRIBUTING.md
new file mode 100644
index 0000000..875ae87
--- /dev/null
+++ b/.github/CONTRIBUTING.md
@@ -0,0 +1,288 @@
+# Contributing to SCWorkflow
+
+## Proposing changes with issues
+
+If you want to make a change, it's a good idea to first
+[open an issue](https://code-review.tidyverse.org/issues/)
+and make sure someone from the team agrees that it’s needed.
+
+If you've decided to work on an issue,
+[assign yourself to the issue](https://docs.github.com/en/issues/tracking-your-work-with-issues/assigning-issues-and-pull-requests-to-other-github-users#assigning-an-individual-issue-or-pull-request)
+so others will know you're working on it.
+
+## Pull request process
+
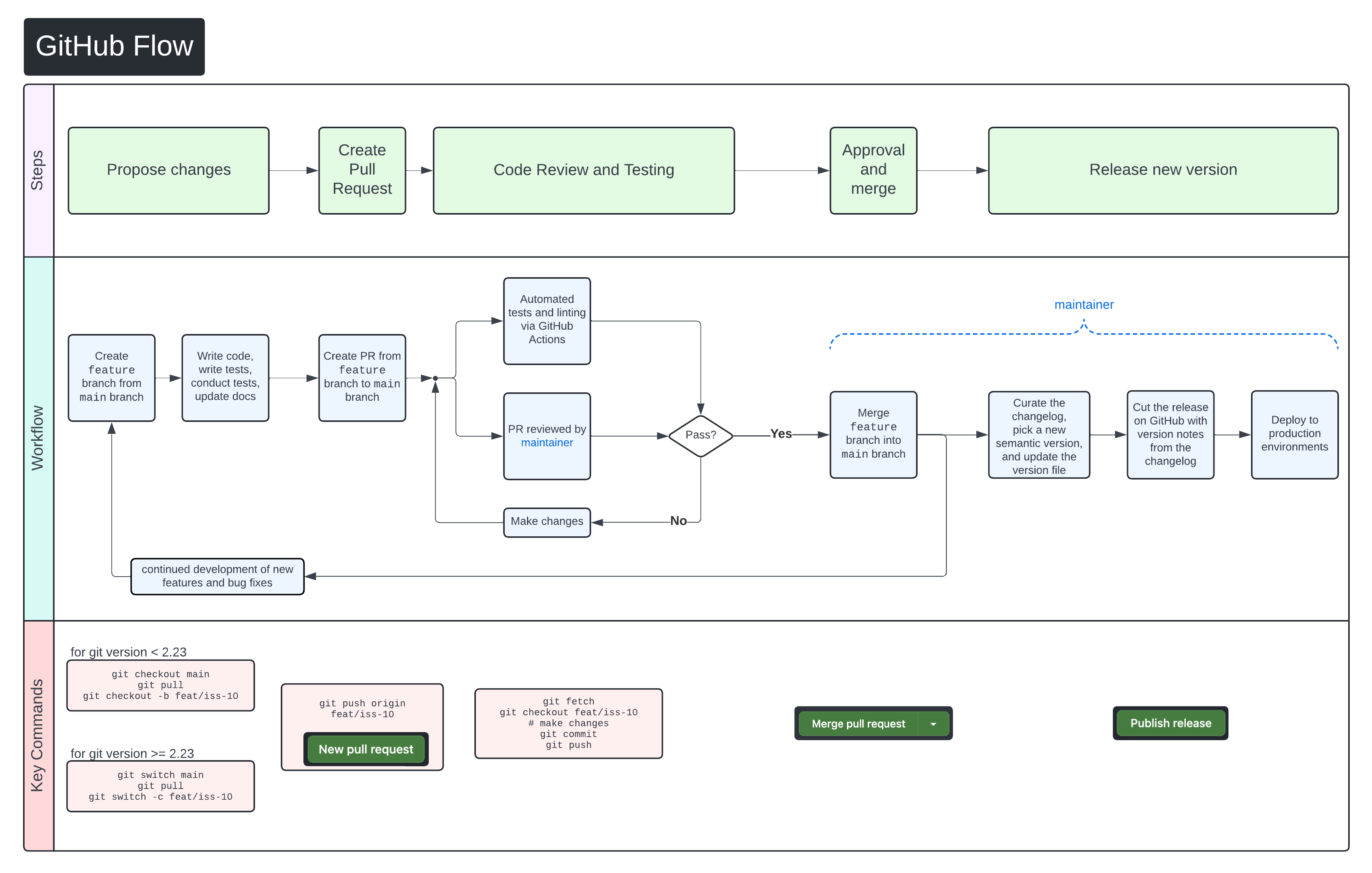
+We use [GitHub Flow](https://docs.github.com/en/get-started/using-github/github-flow)
+as our collaboration process.
+Follow the steps below for detailed instructions on contributing changes to
+SCWorkflow.
+
+
+
+
+### Clone the repo
+
+If you are a member of [CCBR](https://github.com/CCBR),
+you can clone this repository to your computer or development environment.
+Otherwise, you will first need to
+[fork](https://docs.github.com/en/pull-requests/collaborating-with-pull-requests/working-with-forks/fork-a-repo)
+the repo and clone your fork. You only need to do this step once.
+
+```sh
+git clone https://github.com/CCBR/SCWorkflow
+```
+
+> Cloning into 'SCWorkflow'...
+> remote: Enumerating objects: 1136, done.
+> remote: Counting objects: 100% (463/463), done.
+> remote: Compressing objects: 100% (357/357), done.
+> remote: Total 1136 (delta 149), reused 332 (delta 103), pack-reused 673
+> Receiving objects: 100% (1136/1136), 11.01 MiB | 9.76 MiB/s, done.
+> Resolving deltas: 100% (530/530), done.
+
+```sh
+cd SCWorkflow
+```
+
+### If this is your first time cloning the repo, install dependencies
+
+- In an R console, install the R development dependencies with
+ `devtools::install_dev_deps()`, and then make sure the package passes R CMD
+ check by running `devtools::check()`. If R CMD check doesn't pass cleanly,
+ it's a good idea to ask for help before continuing.
+
+- Install [`pre-commit`](https://pre-commit.com/#install) if you don't already
+ have it. Then from the repo's root directory, run
+
+ ```sh
+ pre-commit install
+ ```
+
+ This will install the repo's pre-commit hooks.
+ You'll only need to do this step the first time you clone the repo.
+
+### Create a branch
+
+ Create a Git branch for your pull request (PR). Give the branch a descriptive
+ name for the changes you will make, such as `iss-10` if it is for a specific
+ issue.
+
+ ```sh
+ # create a new branch and switch to it
+ git branch iss-10
+ git switch iss-10
+ ```
+
+ > Switched to a new branch 'iss-10'
+
+### Make your changes
+
+Edit the code, write unit tests, and update the documentation as needed.
+
+#### style
+
+New code should follow the [tidyverse style guide](https://style.tidyverse.org).
+You can use the [styler](https://CRAN.R-project.org/package=styler) package to
+apply these styles, but please don't restyle code that has nothing to do with
+your PR.
+
+A brief overview of conventions according to the tidyverse style guide:
+
+- most object names (variables and functions) should be in [snake_case](https://style.tidyverse.org/syntax.html#sec-objectnames)
+- function names should use [verbs](https://style.tidyverse.org/functions.html#naming) where possible
+- use `<-` for assignment
+- use [pipes](https://style.tidyverse.org/pipes.html) to chain operations on a single object
+
+Please see the [tidyverse style guide](https://style.tidyverse.org) for more details.
+
+#### test
+
+Most changes to the code will also need unit tests to demonstrate that the
+changes work as intended.
+Use [`testthat`](https://testthat.r-lib.org/) to create your unit tests and test
+the code.
+Test files are organized as described in
+.
+Take a look at the existing code in this package for examples.
+
+#### document
+
+If you have written a new function or changed the API of an existing function,
+you will need to update the function's documentation using
+[roxygen2](https://cran.r-project.org/package=roxygen2) with
+[Markdown syntax](https://roxygen2.r-lib.org/articles/rd-formatting.html).
+See instructions on writing roxygen2 comments here:
+.
+If the function is used in a vignette, you may also need to update the vignette.
+
+#### check
+
+After making your changes, run `devtools::check()` from an R console to make
+sure the package still passes R CMD check.
+
+### Commit and push your changes
+
+If you're not sure how often you should commit or what your commits should
+consist of, we recommend following the "atomic commits" principle where each
+commit contains one new feature, fix, or task.
+Learn more about atomic commits here:
+
+
+First, add the files that you changed to the staging area:
+
+```sh
+git add path/to/changed/files/
+```
+
+Then make the commit.
+Your commit message should follow the
+[Conventional Commits](https://www.conventionalcommits.org/en/v1.0.0/)
+specification.
+Briefly, each commit should start with one of the approved types such as
+`feat`, `fix`, `docs`, etc. followed by a description of the commit.
+Take a look at the [Conventional Commits specification](https://www.conventionalcommits.org/en/v1.0.0/#summary)
+for more detailed information about how to write commit messages.
+
+
+```sh
+git commit -m 'feat: create function for awesome feature'
+```
+
+pre-commit will enforce that your commit message and the code changes are
+styled correctly and will attempt to make corrections if needed.
+
+> Check for added large files..............................................Passed
+> Fix End of Files.........................................................Passed
+> Trim Trailing Whitespace.................................................Failed
+> - hook id: trailing-whitespace
+> - exit code: 1
+> - files were modified by this hook
+>
+> Fixing path/to/changed/files/file.txt
+>
+> codespell................................................................Passed
+> style-files..........................................(no files to check)Skipped
+> readme-rmd-rendered..................................(no files to check)Skipped
+> use-tidy-description.................................(no files to check)Skipped
+
+In the example above, one of the hooks modified a file in the proposed commit,
+so the pre-commit check failed. You can run `git diff` to see the changes that
+pre-commit made and `git status` to see which files were modified. To proceed
+with the commit, re-add the modified file(s) and re-run the commit command:
+
+```sh
+git add path/to/changed/files/file.txt
+git commit -m 'feat: create function for awesome feature'
+```
+
+This time, all the hooks either passed or were skipped
+(e.g. hooks that only run on R code will not run if no R files were
+committed).
+When the pre-commit check is successful, the usual commit success message
+will appear after the pre-commit messages showing that the commit was created.
+
+> Check for added large files..............................................Passed
+> Fix End of Files.........................................................Passed
+> Trim Trailing Whitespace.................................................Passed
+> codespell................................................................Passed
+> style-files..........................................(no files to check)Skipped
+> readme-rmd-rendered..................................(no files to check)Skipped
+> use-tidy-description.................................(no files to check)Skipped
+> Conventional Commit......................................................Passed
+> [iss-10 9ff256e] feat: create function for awesome feature
+> 1 file changed, 22 insertions(+), 3 deletions(-)
+
+Finally, push your changes to GitHub:
+
+```sh
+git push
+```
+
+If this is the first time you are pushing this branch, you may have to
+explicitly set the upstream branch:
+
+```sh
+git push --set-upstream origin iss-10
+```
+
+> Enumerating objects: 7, done.
+> Counting objects: 100% (7/7), done.
+> Delta compression using up to 10 threads
+> Compressing objects: 100% (4/4), done.
+> Writing objects: 100% (4/4), 648 bytes | 648.00 KiB/s, done.
+> Total 4 (delta 3), reused 0 (delta 0), pack-reused 0
+> remote: Resolving deltas: 100% (3/3), completed with 3 local objects.
+> remote:
+> remote: Create a pull request for 'iss-10' on GitHub by visiting:
+> remote: https://github.com/CCBR/SCWorkflow/pull/new/iss-10
+> remote:
+> To https://github.com/CCBR/SCWorkflow
+>
+> [new branch] iss-10 -> iss-10
+> branch 'iss-10' set up to track 'origin/iss-10'.
+
+We recommend pushing your commits often so they will be backed up on GitHub.
+You can view the files in your branch on GitHub at
+`https://github.com/CCBR/SCWorkflow/tree/`
+(replace `` with the actual name of your branch).
+
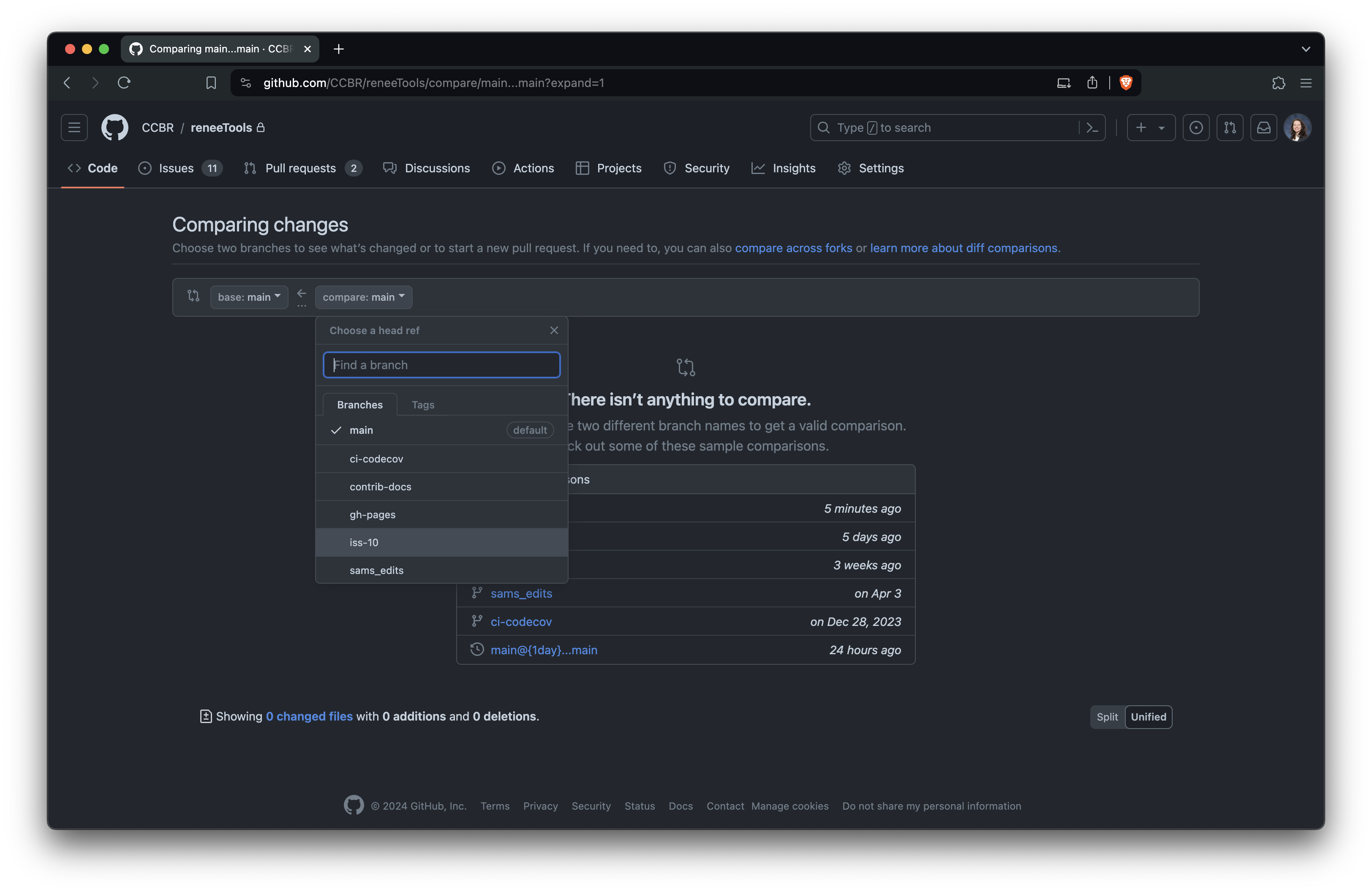
+### Create the PR
+
+Once your branch is ready, create a PR on GitHub:
+
+
+Select the branch you just pushed:
+
+
+
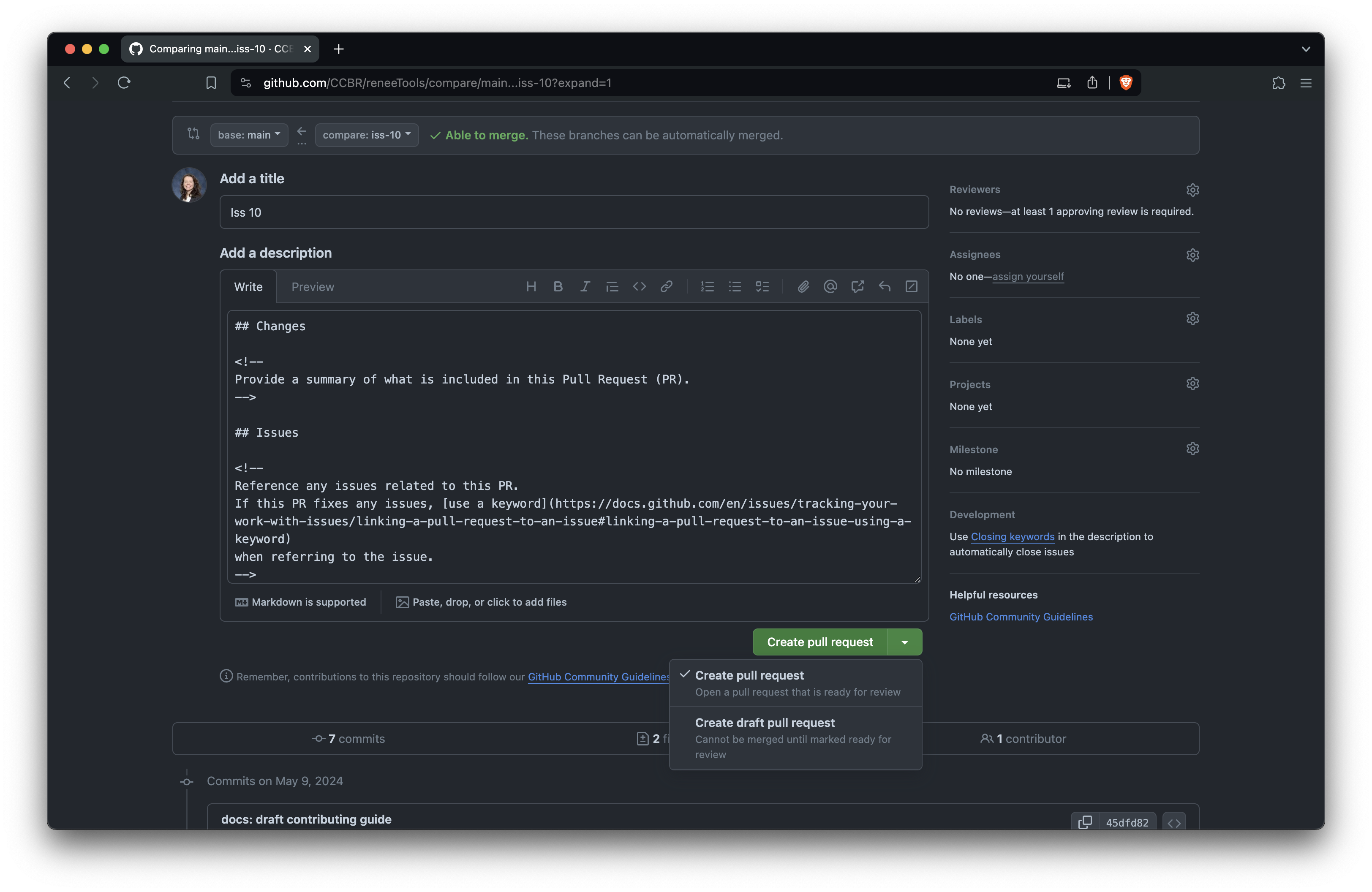
+Edit the PR title and description.
+The title should briefly describe the change.
+Follow the comments in the template to fill out the body of the PR, and
+you can delete the comments (everything between ``) as you go.
+When you're ready, click 'Create pull request' to open it.
+
+
+
+Optionally, you can mark the PR as a draft if you're not yet ready for it to
+be reviewed, then change it later when you're ready.
+
+### Wait for a maintainer to review your PR
+
+We will do our best to follow the tidyverse code review principles:
+.
+The reviewer may suggest that you make changes before accepting your PR in
+order to improve the code quality or style.
+If that's the case, continue to make changes in your branch and push them to
+GitHub, and they will appear in the PR.
+
+Once the PR is approved, the maintainer will merge it and the issue(s) the PR
+links will close automatically.
+Congratulations and thank you for your contribution!
+
+### After your PR has been merged
+
+After your PR has been merged, update your local clone of the repo by
+switching to the main branch and pulling the latest changes:
+
+```sh
+git checkout main
+git pull
+```
+
+It's a good idea to run `git pull` before creating a new branch so it will
+start from the most recent commits in main.
+
+## Helpful links for more information
+
+- This contributing guide was adapted from the [tidyverse contributing guide](https://github.com/tidyverse/tidyverse/blob/main/.github/CONTRIBUTING.md)
+- [GitHub Flow](https://docs.github.com/en/get-started/using-github/github-flow)
+- [tidyverse style guide](https://style.tidyverse.org)
+- [tidyverse code review principles](https://code-review.tidyverse.org)
+- [reproducible examples](https://www.tidyverse.org/help/#reprex)
+- [R packages book](https://r-pkgs.org/)
+- packages:
+ - [usethis](https://usethis.r-lib.org/)
+ - [devtools](https://devtools.r-lib.org/)
+ - [testthat](https://testthat.r-lib.org/)
+ - [styler](https://styler.r-lib.org/)
+ - [roxygen2](https://roxygen2.r-lib.org)
diff --git a/.github/ISSUE_TEMPLATE/bug_report.yml b/.github/ISSUE_TEMPLATE/bug_report.yml
new file mode 100644
index 0000000..95c98d1
--- /dev/null
+++ b/.github/ISSUE_TEMPLATE/bug_report.yml
@@ -0,0 +1,45 @@
+name: Bug report
+description: Report something that is broken or incorrect
+labels: bug
+body:
+ - type: markdown
+ attributes:
+ value: |
+ Before you submit this issue, please check the documentation:
+
+ - type: textarea
+ id: description
+ attributes:
+ label: Description of the bug
+ description: A clear and concise description of what the bug is.
+ validations:
+ required: true
+
+ - type: textarea
+ id: reprex
+ attributes:
+ label: Code and output
+ description: Please include a minimal reproducible example (AKA a reprex). If you've never heard of a [reprex](http://reprex.tidyverse.org/) before, start by reading .
+ render: console
+ placeholder: |
+ library(SCWorkflow)
+ ... insert_your_code_here() ...
+
+ Paste some output where something broke
+
+ - type: textarea
+ id: files
+ attributes:
+ label: Relevant files
+ description: |
+ Please drag and drop any relevant files here if applicable. Create a `.zip` archive if the extension is not allowed.
+
+ - type: textarea
+ id: system
+ attributes:
+ label: System information
+ description: |
+ * Version of R
+ * Version of CCBR/SCWorkflow
+ * OS _(eg. Ubuntu Linux, macOS)_
+ * Hardware _(eg. HPC, Desktop)_
diff --git a/.github/ISSUE_TEMPLATE/config.yml b/.github/ISSUE_TEMPLATE/config.yml
new file mode 100644
index 0000000..84f8757
--- /dev/null
+++ b/.github/ISSUE_TEMPLATE/config.yml
@@ -0,0 +1,4 @@
+contact_links:
+ - name: Discussions
+ url: https://github.com/CCBR/SCWorkflow/discussions
+ about: Please ask and answer questions here.
diff --git a/.github/ISSUE_TEMPLATE/feature_request.yml b/.github/ISSUE_TEMPLATE/feature_request.yml
new file mode 100644
index 0000000..73a08f5
--- /dev/null
+++ b/.github/ISSUE_TEMPLATE/feature_request.yml
@@ -0,0 +1,11 @@
+name: Feature request
+description: Suggest an idea for the package
+labels: enhancement
+body:
+ - type: textarea
+ id: description
+ attributes:
+ label: Description of feature
+ description: Please describe your suggestion for a new feature. It might help to describe a problem or use case, plus any alternatives that you have considered.
+ validations:
+ required: true
diff --git a/.github/PULL_REQUEST_TEMPLATE.md b/.github/PULL_REQUEST_TEMPLATE.md
new file mode 100644
index 0000000..bc679fa
--- /dev/null
+++ b/.github/PULL_REQUEST_TEMPLATE.md
@@ -0,0 +1,24 @@
+## Changes
+
+
+
+## Issues
+
+
+
+## PR Checklist
+
+(~Strikethrough~ any points that are not applicable.)
+
+- [ ] This comment contains a description of changes with justifications, with any relevant issues linked.
+- [ ] Write unit tests for any new features, bug fixes, or other code changes.
+- [ ] Update the docs if there are any API changes (roxygen2 comments, vignettes, readme, etc.).
+- [ ] Update `NEWS.md` with a short description of any user-facing changes and reference the PR number. Follow the style described in
+- [ ] Run `devtools::check()` locally and fix all notes, warnings, and errors.
diff --git a/.github/package-versions.txt b/.github/package-versions.txt
new file mode 100644
index 0000000..2facfd9
--- /dev/null
+++ b/.github/package-versions.txt
@@ -0,0 +1 @@
+any::Seurat@4.1.1, any::Matrix@1.5.1
\ No newline at end of file
diff --git a/.github/workflows/R-CMD-check.yaml b/.github/workflows/R-CMD-check.yaml
new file mode 100644
index 0000000..9a97fff
--- /dev/null
+++ b/.github/workflows/R-CMD-check.yaml
@@ -0,0 +1,85 @@
+# Workflow derived from https://github.com/r-lib/actions/tree/v2/examples
+# Need help debugging build failures? Start at https://github.com/r-lib/actions#where-to-find-help
+on:
+ push:
+ branches: [main, master, dev, DEV]
+ pull_request:
+ branches: [main, master, dev, DEV]
+ workflow_dispatch:
+
+name: R-CMD-check
+
+permissions:
+ contents: read
+ pull-requests: read
+
+jobs:
+ R-CMD-check:
+
+ strategy:
+ fail-fast: false
+ matrix:
+ config:
+ - { os: ubuntu-latest, r: '4.1.3' }
+ #- { os: ubuntu-latest, r: 'oldrel-1' }
+ #- { os: macos-latest, r: 'release' }
+
+ runs-on: ${{ matrix.config.os }}
+ name: ${{ matrix.config.os }} (${{ matrix.config.r }})
+ env:
+ GITHUB_PAT: ${{ secrets.GITHUB_TOKEN }}
+ R_KEEP_PKG_SOURCE: yes
+
+ steps:
+ - uses: actions/checkout@v4
+
+ - uses: CCBR/actions/install-r-pak@main
+ with:
+ versions-file: .github/package-versions.txt
+ extra-packages: local::.
+ needs: dev
+ r-version: ${{ matrix.config.r }}
+ http-user-agent: ${{ matrix.config.http-user-agent }}
+
+ - uses: r-lib/actions/check-r-package@v2
+ with:
+ upload-snapshots: true
+
+ lint:
+ runs-on: ubuntu-latest
+ steps:
+ - uses: actions/checkout@v4
+
+ - uses: CCBR/actions/install-r-pak@main
+ with:
+ r-version: 4.1.3
+ versions-file: .github/package-versions.txt
+ needs: dev
+
+ - name: Good Practice checks
+ shell: Rscript {0}
+ run: |
+ g <- goodpractice::gp()
+ g
+ n_failed <- length(goodpractice::failed_checks(g))
+ if (n_failed > 0) {
+ warning(paste(n_failed, "failed checks"))
+ }
+
+ - name: Lint
+ shell: Rscript {0}
+ run: lintr::lint_package()
+ env:
+ LINTR_ERROR_ON_LINT: false
+
+ check: # make sure all check jobs pass. https://github.com/orgs/community/discussions/4324#discussioncomment-3477871
+ runs-on: ubuntu-latest
+ needs: [R-CMD-check, lint]
+ if: always()
+ steps:
+ - name: Successful build
+ if: ${{ !(contains(needs.*.result, 'failure')) }}
+ run: exit 0
+ - name: Failing build
+ if: ${{ contains(needs.*.result, 'failure') }}
+ run: exit 1
diff --git a/.github/workflows/pkgdown.yaml b/.github/workflows/pkgdown.yaml
index f9745ba..0459bb7 100644
--- a/.github/workflows/pkgdown.yaml
+++ b/.github/workflows/pkgdown.yaml
@@ -2,7 +2,7 @@
# Need help debugging build failures? Start at https://github.com/r-lib/actions#where-to-find-help
on:
push:
- branches: [main, master, GalaxyCLI]
+ branches: [main, master]
pull_request:
branches: [main, master]
release:
@@ -26,17 +26,12 @@ jobs:
steps:
- uses: actions/checkout@v3
- - uses: r-lib/actions/setup-pandoc@v2
-
- - uses: r-lib/actions/setup-r@v2
- with:
- use-public-rspm: true
- r-version: 4.3
-
- - uses: r-lib/actions/setup-r-dependencies@v2
+ - uses: CCBR/actions/install-r-pak@main
with:
- extra-packages: any::pkgdown, local::.
- needs: website
+ r-version: 4.1.3
+ versions-file: .github/package-versions.txt
+ extra-packages: local::.
+ needs: dev
- name: Build site
run: pkgdown::build_site_github_pages(new_process = FALSE, install = FALSE)
diff --git a/.github/workflows/test-coverage.yaml b/.github/workflows/test-coverage.yaml
new file mode 100644
index 0000000..4ac1c43
--- /dev/null
+++ b/.github/workflows/test-coverage.yaml
@@ -0,0 +1,58 @@
+# Workflow derived from https://github.com/r-lib/actions/tree/v2/examples
+# Need help debugging build failures? Start at https://github.com/r-lib/actions#where-to-find-help
+on:
+ push:
+ branches: [main, master, dev, DEV]
+ pull_request:
+ branches: [main, master, dev, DEV]
+
+name: test-coverage
+
+permissions: read-all
+
+jobs:
+ test-coverage:
+ runs-on: ubuntu-latest
+ env:
+ GITHUB_PAT: ${{ secrets.GITHUB_TOKEN }}
+
+ steps:
+ - uses: actions/checkout@v4
+
+ - uses: CCBR/actions/install-r-pak@main
+ with:
+ r-version: 4.1.3
+ versions-file: .github/package-versions.txt
+ needs: dev
+
+ - name: Test coverage
+ run: |
+ cov <- covr::package_coverage(
+ quiet = FALSE,
+ clean = FALSE,
+ install_path = file.path(normalizePath(Sys.getenv("RUNNER_TEMP"), winslash = "/"), "package")
+ )
+ covr::to_cobertura(cov)
+ shell: Rscript {0}
+
+ - uses: codecov/codecov-action@v4
+ with:
+ fail_ci_if_error: ${{ github.event_name != 'pull_request' && true || false }}
+ file: ./cobertura.xml
+ plugin: noop
+ disable_search: true
+ token: ${{ secrets.CODECOV_TOKEN }}
+
+ - name: Show testthat output
+ if: always()
+ run: |
+ ## --------------------------------------------------------------------
+ find '${{ runner.temp }}/package' -name 'testthat.Rout*' -exec cat '{}' \; || true
+ shell: bash
+
+ - name: Upload test results
+ if: failure()
+ uses: actions/upload-artifact@v4
+ with:
+ name: coverage-test-failures
+ path: ${{ runner.temp }}/package
diff --git a/.github/workflows/user-projects.yml b/.github/workflows/user-projects.yml
new file mode 100644
index 0000000..c189ef6
--- /dev/null
+++ b/.github/workflows/user-projects.yml
@@ -0,0 +1,23 @@
+name: user-projects
+
+on:
+ issues:
+ types:
+ - assigned
+ pull_request:
+ types:
+ - assigned
+
+permissions:
+ issues: write
+ pull-requests: write
+
+jobs:
+ add-to-project:
+ runs-on: ubuntu-latest
+ steps:
+ - uses: CCBR/actions/user-projects@main
+ with:
+ app-id: ${{ vars.CCBR_BOT_APP_ID }}
+ app-private-key: ${{ secrets.CCBR_BOT_PRIVATE_KEY }}
+ token-owner: "CCBR"
diff --git a/.gitignore b/.gitignore
index 55ed9b9..17be9e8 100755
--- a/.gitignore
+++ b/.gitignore
@@ -5,4 +5,3 @@ Rcheck.txt
*.pdf
tests/testthat/otherData/
.Rproj.user
-*.txt
diff --git a/DESCRIPTION b/DESCRIPTION
index cdf6c35..3bbcff8 100755
--- a/DESCRIPTION
+++ b/DESCRIPTION
@@ -1,89 +1,103 @@
Package: SCWorkflow
Title: SCWorkflow from NIDAP
Version: 1.0.2
-Authors@R: c(person("Maggie", "Cam", email = "maggie.cam@nih.gov", role = "aut", comment = c(ORCID = "0000-0001-8190-9766")),
- person("Thomas", "Meyer", email = "thomas.meyer@nih.gov", role = c("aut", "cre"), comment = c(ORCID = "0000-0002-7185-5597")),
- person("Jing", "Bian", email = "bianjh@nih.gov", role = "aut", comment = c(ORCID = "0000-0001-7109-716X")),
- person("Alexandra", "Michalowski", email = "michaloa@mail.nih.gov", role = "aut", comment = c(ORCID = "0000-0001-9259-6101")),
- person("Alexei", "Lobanov", email = "alexei.lobanov@nih.gov", role = "aut", comment = c(ORCID = "0000-0002-9883-4374")),
- person("Philip", "Homan", email = "philip.homan@nih.gov", role = "aut", comment = c(ORCID = "0000-0002-3389-4931")),
- person("Rui", "He", email = "rui.he@nih.gov", role = "aut"))
-Description: A set of functions for analyzing single-cell RNA-seq data using the
- Seurat workflow. The user provides H5 files containing the results of the
- upstream processing through CellRanger, and the package functions allow for
- the QC, filtering, normalization, annotation, differential gene expression,
- and further visualizations and analysis based on user input. This package can
- be run both in a docker container and in user-friendly web-based interactive
- notebooks (NIDAP, Palantir Foundry).
+Authors@R: c(
+ person("Maggie", "Cam", , "maggie.cam@nih.gov", role = "aut",
+ comment = c(ORCID = "0000-0001-8190-9766")),
+ person("Thomas", "Meyer", , "thomas.meyer@nih.gov", role = c("aut", "cre"),
+ comment = c(ORCID = "0000-0002-7185-5597")),
+ person("Jing", "Bian", , "bianjh@nih.gov", role = "aut",
+ comment = c(ORCID = "0000-0001-7109-716X")),
+ person("Alexandra", "Michalowski", , "michaloa@mail.nih.gov", role = "aut",
+ comment = c(ORCID = "0000-0001-9259-6101")),
+ person("Alexei", "Lobanov", , "alexei.lobanov@nih.gov", role = "aut",
+ comment = c(ORCID = "0000-0002-9883-4374")),
+ person("Philip", "Homan", , "philip.homan@nih.gov", role = "aut",
+ comment = c(ORCID = "0000-0002-3389-4931")),
+ person("Rui", "He", , "rui.he@nih.gov", role = "aut")
+ )
+Description: A set of functions for analyzing single-cell RNA-seq data
+ using the Seurat workflow. The user provides H5 files containing the
+ results of the upstream processing through CellRanger, and the package
+ functions allow for the QC, filtering, normalization, annotation,
+ differential gene expression, and further visualizations and analysis
+ based on user input. This package can be run both in a docker
+ container and in user-friendly web-based interactive notebooks (NIDAP,
+ Palantir Foundry).
License: MIT
-Encoding: UTF-8
-Roxygen: list(markdown = TRUE)
-RoxygenNote: 7.2.3
-Suggests:
- testthat (>= 3.0.0)
Depends:
R (>= 4.0)
Imports:
anndata (>= 0.7.5.2),
+ BiocManager,
callr (>= 3.7.1),
+ celldex,
+ colorspace,
+ ComplexHeatmap (>= 2.10.0),
cowplot (>= 1.1.1),
data.table (>= 1.14.2),
+ dendextend,
+ dendsort,
+ digest (>= 0.6.29),
dplyr (>= 1.0.9),
edgeR (>= 3.36.0),
- future.apply (>= 1.9.0),
future (>= 1.27.0),
+ future.apply (>= 1.9.0),
gargle (>= 1.2.0),
+ gdata,
+ ggExtra,
ggplot2 (>= 3.3.6),
ggpubr (>= 0.4.0),
+ ggrepel,
globals (>= 0.16.1),
+ gridBase (>= 0.4-7),
+ gridExtra (>= 2.3),
+ gtable (>= 0.3.1),
harmony (>= 0.1.1),
hdf5r (>= 1.3.5),
+ htmlwidgets,
httpuv (>= 1.6.5),
+ httr,
+ jsonlite,
leiden (>= 0.4.2),
limma (>= 3.50.3),
magrittr (>= 2.0.3),
markdown (>= 1.1),
+ MAST (>= 1.20.0),
methods (>= 4.1.3),
- plotly (>= 4.10.0),
+ pheatmap,
+ plotly (>= 4.10.0),
+ plyr,
+ png,
progressr (>= 0.10.1),
- pryr (>= 0.1.5),
purrr (>= 0.3.4),
quantmod (>= 0.4.20),
+ RColorBrewer (>= 1.1-3),
reshape2 (>= 1.4.4),
reticulate (>= 1.25),
rlang (>= 1.0.6),
+ scales,
+ scDblFinder,
+ Seurat (>= 4.1.1),
+ SingleR (>= 1.8.1),
statmod (>= 1.4.37),
stringr (>= 1.4.1),
svglite (>= 2.1.0),
tibble (>= 3.1.8),
+ tidyr,
tidyverse (>= 1.3.2),
viridisLite (>= 0.4.0),
xfun (>= 0.32),
- zip (>= 2.2.0),
- ComplexHeatmap (>= 2.10.0),
- MAST (>= 1.20.0),
- SingleR (>= 1.8.1),
- BiocManager,
- gridBase (>= 0.4-7),
- gridExtra (>= 2.3),
- RColorBrewer (>= 1.1-3),
- Seurat (>= 4.1.1),
- gtable (>= 0.3.1),
- digest (>= 0.6.29),
- png,
- ggExtra,
- httr,
- jsonlite,
- plyr,
- colorspace,
- dendextend,
- dendsort,
- pheatmap,
- scales,
- celldex,
- gdata,
- ggrepel,
- tidyr,
- htmlwidgets,
- scDblFinder
+ zip (>= 2.2.0)
+Suggests:
+ knitr,
+ rmarkdown,
+ roxygen2,
+ testthat (>= 3.0.0),
+ usethis
+Config/Needs/dev: cffr, covr, goodpractice, here, lintr, pkgdown,
+ rcmdcheck
Config/testthat/edition: 3
+Encoding: UTF-8
+Roxygen: list(markdown = TRUE)
+RoxygenNote: 7.2.3
diff --git a/Dockerfile b/Dockerfile
index b91c92f..e957f2a 100644
--- a/Dockerfile
+++ b/Dockerfile
@@ -1,64 +1,548 @@
-FROM nciccbr/ccbr_ubuntu_22.04:v4
-
-# build time variables
-ARG BUILD_DATE="000000"
-ENV BUILD_DATE=${BUILD_DATE}
-ARG BUILD_TAG="000000"
-ENV BUILD_TAG=${BUILD_TAG}
-ARG REPONAME="000000"
-ENV REPONAME=${REPONAME}
-
-ARG R_VERSION=4.3.2
-ENV R_VERSION=${R_VERSION}
-
-SHELL ["/bin/bash", "-lc"]
-
-# Install conda and give write permissions to conda folder
-RUN echo 'export PATH=/opt2/conda/bin:$PATH' > /etc/profile.d/conda.sh && \
- wget --quiet "https://github.com/conda-forge/miniforge/releases/latest/download/Miniforge3-$(uname)-$(uname -m).sh" -O ~/miniforge3.sh && \
- /bin/bash ~/miniforge3.sh -b -p /opt2/conda && \
- rm ~/miniforge3.sh && chmod 777 -R /opt2/conda/
-ENV PATH="/opt2/conda/bin:$PATH"
-
-# Pin channels and update
-RUN conda config --add channels conda-forge \
- && conda config --add channels bioconda \
- && conda config --set channel_priority strict
-
-# install conda packages
-RUN mamba install -y -c conda-forge \
- r-base=${R_VERSION} \
- r-devtools \
- r-ggplot2 \
- r-ggrepel r-viridis r-upsetr r-patchwork r-plotly \
- r-matrix r-mgcv r-survival \
- bioconductor-genomicranges \
- bioconductor-summarizedexperiment \
- bioconductor-delayedarray \
- bioconductor-s4arrays \
- bioconductor-annotationdbi \
- bioconductor-annotate \
- bioconductor-keggrest \
- && conda clean -afy
-
-# install R package
-COPY . /opt2/SCWorkflow
-RUN R -e "devtools::install_local('/opt2/SCWorkflow', dependencies = TRUE, repos='http://cran.rstudio.com')"
-
-# add scworkflow exec to the path
-# RUN chmod -R +x /opt2/conda/lib/R/library/SCWorkflow/exec
-# ENV PATH="$PATH:/opt2/conda/lib/R/library/SCWorkflow/exec"
-# RUN scworkflow --help
-
-# copy example script & json to data
-COPY ./inst/extdata/example_script.sh /data2/
-COPY ./inst/extdata/json_args/ /data2/json_args/
-
-# Save Dockerfile in the docker
-COPY Dockerfile /opt2/Dockerfile_${REPONAME}.${BUILD_TAG}
-RUN chmod a+r /opt2/Dockerfile_${REPONAME}.${BUILD_TAG}
-
-# cleanup
-WORKDIR /data2
-RUN apt-get clean && apt-get purge \
- && rm -rf /var/lib/apt/lists/* /tmp/* /var/tmp/*
+FROM rocker/rstudio:4.1.3
+
+RUN apt-get update && apt-get install -y --no-install-recommends \
+ bash gcc g++ gfortran make cmake pkg-config \
+ git vim-tiny \
+ libcurl4-openssl-dev libssl-dev libxml2-dev \
+ libpng-tools \
+ libpng-dev libjpeg-dev libtiff5-dev zlib1g-dev \
+ libfreetype6-dev libharfbuzz-dev libfribidi-dev \
+ libbz2-dev liblzma-dev \
+ libgsl-dev libhdf5-dev \
+ tcl tk tcl-dev tk-dev \
+ libglpk-dev libglpk40 \
+ libgit2-dev \
+ python3 python3-venv \
+ python3-dev python3-pip \
+ libfontconfig1-dev \
+ autoconf automake libtool \
+ libgeos-dev libproj-dev \
+ libcairo2-dev libxt-dev \
+ automake \
+ && rm -rf /var/lib/apt/lists/*
+
+
+# libpng libtiff-4 libjpeg libwebp libwebpmux
+
+# (Optional) prevent R networking during local install steps
+# xxx rpf get rid of
+# RUN echo 'options(repos = c())' >> /usr/local/lib/R/etc/Rprofile.site
+
+#RUN pip3 install --no-cache-dir igraph leidenalg numpy
+# weird pip instal bug introduced
+RUN pip3 install --no-cache-dir igraph "leidenalg==0.10.0" numpy
+
+WORKDIR /home/rstudio
+
+# Remove any spatstat* that might be preinstalled in the rocker image
+RUN R -q -e " \
+ for (lib in .libPaths()) { \
+ ip <- rownames(installed.packages(lib.loc = lib)); \
+ pk <- grep('^spatstat', ip, value = TRUE); \
+ if (length(pk)) { \
+ message('Removing from ', lib, ': ', paste(pk, collapse=', ')); \
+ remove.packages(pk, lib = lib); \
+ } \
+ }"
+
+# Install packages with specific versions from CRAN and Bioconductor
+RUN R --vanilla --slave << 'EOF'
+options(repos = c(CRAN = 'https://cran.r-project.org'))
+
+if (!require('BiocManager', quietly = TRUE)) {
+ install.packages('BiocManager', quiet = TRUE)
+}
+if (!require('remotes', quietly = TRUE)) {
+ install.packages('remotes', quiet = TRUE)
+}
+
+# Install specific CRAN package versions
+cran_versions <- list(
+ bitops = '1.0-8',
+ RCurl = '1.98-1.14',
+ GlobalOptions = '0.1.2',
+ bit = '4.0.4',
+ assertthat = '0.2.1',
+ bit64 = '4.0.5',
+ shape = '1.4.6',
+ colorspace = '2.0-3',
+ circlize = '0.4.15',
+ rlang = '1.1.4',
+ cli = '3.6.3',
+ glue = '1.7.0',
+ lifecycle = '1.0.4',
+ vctrs = '0.6.5',
+ blob = '1.2.3',
+ clue = '0.3-61',
+ cluster = '2.1.2',
+ codetools = '0.2-18',
+ crayon = '1.5.3',
+ data.table = '1.15.4',
+ DBI = '1.2.3',
+ dendsort = '0.3.4',
+ digest = '0.6.37',
+ ellipsis = '0.3.2',
+ evaluate = '0.24.0',
+ fansi = '1.0.6',
+ farver = '2.1.1',
+ fastmap = '1.2.0',
+ cachem = '1.1.0',
+ fastmatch = '1.1-3',
+ iterators = '1.0.14',
+ foreach = '1.5.2',
+ formatR = '1.14',
+ generics = '0.1.3',
+ rjson = '0.2.21',
+ GetoptLong = '1.0.5',
+ gtable = '0.3.5',
+ labeling = '0.4.2',
+ munsell = '0.5.0',
+ R6 = '2.5.1',
+ gridExtra = '2.3',
+ RColorBrewer = '1.1-3',
+ utf8 = '1.2.4',
+ pillar = '1.9.0',
+ pkgconfig = '2.0.3',
+ viridisLite = '0.4.1',
+ scales = '1.2.1',
+ withr = '3.0.1',
+ gtools = '3.9.5',
+ gridGraphics = '0.5-1',
+ hms = '1.1.2',
+ irlba = '2.3.5.1',
+ jsonlite = '1.8.8',
+ png = '0.1-7',
+ KernSmooth = '2.23-20',
+ lambda.r = '1.2.4',
+ lattice = '0.20-45',
+ lazyeval = '0.2.2',
+ locfit = '1.5-9.9',
+ Matrix = '1.5-1',
+ matrixStats = '0.62.0',
+ memoise = '2.0.1',
+ mgcv = '1.8-39',
+ nlme = '3.1-155',
+ rsvd = '1.0.5',
+ Rcpp = '1.0.13',
+ uuid = '1.1-0',
+ xfun = '0.47',
+ xtable = '1.8-4',
+ yaml = '2.3.10',
+ beeswarm = '0.4.0',
+ filelock = '1.0.3',
+ bslib = '0.4.2',
+ Cairo = '1.6-0',
+ desc = '1.4.2',
+ pkgload = '1.3.0',
+ brio = '1.1.3',
+ zip = '2.3.3',
+ openxlsx = '4.2.5.2',
+ praise = '1.0.0',
+ diffobj = '0.3.5',
+ waldo = '0.5.1',
+ testthat = '3.1.6',
+ nloptr = '2.0.3',
+ minqa = '1.2.5',
+ lme4 = '1.1-33',
+ MatrixModels = '0.5-1',
+ SparseM = '1.81',
+ quantreg = '5.95',
+ numDeriv = '2016.8-1.1',
+ pbkrtest = '0.5.2',
+ maptools = '1.1-7',
+ carData = '3.0-5',
+ rio = '0.5.29',
+ car = '3.0-0',
+ pROC = '1.18.2',
+ proxy = '0.4-27',
+ e1071 = '1.7-13',
+ ModelMetrics = '1.2.2.2',
+ clock = '0.7.0',
+ shinyjs = '2.1.0',
+ colourpicker = '1.2.0',
+ combinat = '0.0-8',
+ corrplot = '0.92',
+ cpp11 = '0.4.7',
+ diagram = '1.6.5',
+ DT = '0.28',
+ Rttf2pt1 = '1.3.12',
+ extrafontdb = '1.0',
+ extrafont = '0.18',
+ vipor = '0.4.7',
+ ggbeeswarm = '0.7.2',
+ ggrastr = '1.0.2',
+ fastICA = '1.2-3',
+ gdata = '2.18.0.1',
+ ggExtra = '0.10.1',
+ ggsci = '3.0.0',
+ ggsignif = '0.6.3',
+ gower = '1.0.1',
+ gridBase = '0.4-7',
+ hardhat = '1.3.0',
+ RhpcBLASctl = '0.23-42',
+ hdf5r = '1.3.5',
+ hexbin = '1.28.3',
+ survival = '3.2-13',
+ MASS = '7.3-55',
+ SQUAREM = '2021.1',
+ lava = '1.7.2.1',
+ lobstr = '1.1.2',
+ lsei = '1.3-0',
+ markdown = '1.13',
+ lpSolve = '5.6.16',
+ mclust = '6.0.0',
+ npsurv = '0.5-0',
+ pheatmap = '1.0.12',
+ polynom = '1.4-1',
+ proj4 = '1.0-12',
+ pryr = '0.1.5',
+ ps = '1.8.1',
+ RcppParallel = '5.1.6',
+ rstatix = '0.7.0',
+ timeDate = '4022.108',
+ maps = '3.4.1',
+ ggpubr = '0.4.0',
+ prodlim = '2023.03.31',
+ ipred = '0.9-14',
+ recipes = '1.0.6',
+ caret = '6.0-94',
+ quantmod = '0.4.20',
+ profvis = '0.3.7',
+ sessioninfo = '1.2.3',
+ downlit = '0.4.2',
+ whisker = '0.4.1',
+ pkgdown = '2.0.7',
+ pkgbuild = '1.4.8',
+ brew = '1.0-8',
+ roxygen2 = '7.2.3',
+ urlchecker = '1.0.1',
+ rversions = '2.1.2',
+ xopen = '1.0.0',
+ rcmdcheck = '1.4.0',
+ credentials = '1.3.2',
+ gert = '1.9.2',
+ gitcreds = '0.1.2',
+ httr2 = '0.2.3',
+ ini = '0.3.1',
+ gh = '1.4.0',
+ usethis = '3.1.0',
+ devtools = '2.4.5',
+ ggalt = '0.4.0',
+ EnhancedVolcano = '1.12.0'
+)
+
+for (pkg in names(cran_versions)) {
+ tryCatch({
+ remotes::install_version(pkg, version = cran_versions[[pkg]], repos = 'https://cran.r-project.org', quiet = TRUE)
+ }, error = function(e) {
+ message('Note: install_version failed for ', pkg, ', trying install.packages')
+ install.packages(pkg, quiet = TRUE)
+ })
+}
+
+# Install Bioconductor packages with specific versions
+bioc_versions <- list(
+ BiocGenerics = '0.40.0',
+ Biobase = '2.54.0',
+ S4Vectors = '0.32.4',
+ IRanges = '2.28.0',
+ XVector = '0.34.0',
+ GenomeInfoDbData = '1.2.7',
+ GenomeInfoDb = '1.30.1',
+ beachmat = '2.10.0',
+ edgeR = '3.36.0',
+ GenomicRanges = '1.46.1',
+ Biostrings = '2.62.0',
+ DelayedArray = '0.20.0',
+ sparseMatrixStats = '1.6.0',
+ DelayedMatrixStats = '1.16.0',
+ ScaledMatrix = '1.2.0',
+ zlibbioc = '1.40.0',
+ KEGGREST = '1.34.0',
+ AnnotationDbi = '1.56.2',
+ BiocParallel = '1.28.3',
+ BiocSingular = '1.10.0',
+ ComplexHeatmap = '2.10.0',
+ fgsea = '1.20.0',
+ BiocNeighbors = '1.12.0',
+ BiocFileCache = '2.2.1',
+ bluster = '1.4.0',
+ scuttle = '1.4.0',
+ scater = '1.22.0',
+ scran = '1.22.1',
+ SingleR = '1.8.1',
+ MAST = '1.20.0',
+ scDblFinder = '1.8.0',
+ SummarizedExperiment = '1.24.0',
+ SingleCellExperiment = '1.16.0',
+ HDF5Array = '1.22.1',
+ rhdf5 = '2.38.1',
+ rhdf5filters = '1.6.0',
+ GSVA = '1.42.0',
+ ExperimentHub = '2.2.1',
+ AnnotationHub = '3.2.2',
+ celldex = '1.4.0',
+ annotate = '1.72.0',
+ graph = '1.72.0',
+ GSEABase = '1.56.0',
+ interactiveDisplayBase = '1.32.0',
+ TrajectoryUtils = '1.2.0',
+ TSCAN = '1.32.0',
+ conquer = '1.3.3',
+ metapod = '1.2.0',
+ statmod = '1.5.0'
+)
+
+for (pkg in names(bioc_versions)) {
+ tryCatch({
+ remotes::install_version(pkg, version = bioc_versions[[pkg]], repos = BiocManager::repositories(), quiet = TRUE)
+ }, error = function(e) {
+ message('Note: install_version failed for ', pkg, ', trying BiocManager')
+ BiocManager::install(pkg, ask = FALSE, quiet = TRUE)
+ })
+}
+EOF
+
+
+# Additional packages that need special installation
+
+# Install SCWorkflow from GitHub
+COPY . /opt/SCWorkflow
+RUN R --vanilla --slave -e "remotes::install_local('/opt/SCWorkflow', dependencies = TRUE, quiet = TRUE, upgrade='never')"
+
+# Install spatstat family packages with specific versions
+RUN cat > /tmp/install_spatstat.R << 'EOFSPAT'
+options(repos = c(CRAN = 'https://cran.r-project.org'))
+if (!require('remotes', quietly = TRUE)) install.packages('remotes', quiet = TRUE)
+spatstat_versions <- list(
+ spatstat.utils = '3.1-0',
+ spatstat.data = '3.0-0',
+ deldir = '2.0-4',
+ polyclip = '1.10-7',
+ spatstat.univar = '2.0-3',
+ spatstat.geom = '3.0-3',
+ spatstat.random = '2.2-0',
+ abind = '1.4-5',
+ tensor = '1.5',
+ goftest = '1.2-3',
+ spatstat.sparse = '3.1-0',
+ spatstat.core = '2.4-2',
+ spatstat.linnet = '2.2-1'
+)
+for (pkg in names(spatstat_versions)) {
+ tryCatch({
+ remotes::install_version(pkg, version = spatstat_versions[[pkg]], repos = 'https://cran.r-project.org', quiet = TRUE)
+ }, error = function(e) {
+ message('Note: install_version for ', pkg, ' failed, trying install.packages')
+ install.packages(pkg, quiet = TRUE)
+ })
+}
+EOFSPAT
+
+RUN R --vanilla --slave --file=/tmp/install_spatstat.R
+
+# Install remaining specialized packages
+RUN cat > /tmp/install_special.R << 'EOFSPEC'
+options(repos = c(CRAN = 'https://cran.r-project.org'))
+if (!require('remotes', quietly = TRUE)) install.packages('remotes', quiet = TRUE)
+special_versions <- list(
+ RSpectra = '0.16-1',
+ dotCall64 = '1.1-1',
+ spam = '2.11-0',
+ RcppHNSW = '0.4.1',
+ leidenbase = '0.1.30',
+ fastDummies = '1.7.3',
+ sp = '1.5-0',
+ rgeos = '0.5-9',
+ SeuratObject = '4.1.1',
+ Seurat = '4.1.1',
+ XML = '3.99-0.14',
+ anndata = '0.7.5.2',
+ ash = '1.0-15',
+ viridis = '0.6.5',
+ dendextend = '1.16.0',
+ ggrepel = '0.9.5',
+ l2p = '0.0-13',
+ l2psupp = '0.0-13',
+ ica = '1.0-3',
+ Rtsne = '0.16',
+ ggridges = '0.5.3',
+ scattermore = '1.2',
+ listenv = '0.9.1',
+ globals = '0.16.3',
+ parallelly = '1.38.0',
+ future = '1.34.0',
+ future.apply = '1.11.2',
+ RcppEigen = '0.3.3.9.3',
+ RcppAnnoy = '0.0.19',
+ zoo = '1.8-12',
+ lmtest = '0.9-40',
+ fitdistrplus = '1.1-8',
+ caTools = '1.18.2',
+ gplots = '3.1.3',
+ ROCR = '1.0-11',
+ igraph = '2.0.3',
+ pbapply = '1.5-0',
+ commonmark = '1.9.0',
+ httpuv = '1.6.15',
+ sourcetools = '0.1.7-1',
+ shiny = '1.9.1',
+ miniUI = '0.1.1.1',
+ progressr = '0.14.0',
+ sitmo = '2.0.2',
+ dqrng = '0.4.1',
+ FNN = '1.1.3.2',
+ RcppProgress = '0.4.2',
+ uwot = '0.1.14',
+ cowplot = '1.1.1',
+ RcppTOML = '0.2.2',
+ rprojroot = '2.0.4',
+ here = '1.0.1',
+ reticulate = '1.40.0',
+ leiden = '0.4.3',
+ RANN = '2.6.1',
+ RcppArmadillo = '0.12.4.0.0',
+ sctransform = '0.3.4',
+ Rhdf5lib = '1.16.0',
+ xgboost = '1.7.8.1',
+ nidapFunctions = '0.7.8',
+ snow = '0.4-4',
+ BH = '1.81.0-1',
+ futile.options = '1.0.1',
+ futile.logger = '1.4.3',
+ base64enc = '0.1-3',
+ htmltools = '0.5.8.1',
+ rappdirs = '0.3.3',
+ jquerylib = '0.1.4',
+ tinytex = '0.44',
+ fs = '1.6.4',
+ sass = '0.4.6',
+ mime = '0.12',
+ magrittr = '2.0.3',
+ stringi = '1.8.4',
+ stringr = '1.5.1',
+ highr = '0.10',
+ knitr = '1.48',
+ fontawesome = '0.5.1',
+ rmarkdown = '2.28',
+ htmlwidgets = '1.6.4',
+ sys = '3.4.2',
+ askpass = '1.1',
+ openssl = '2.0.5',
+ curl = '6.4.0',
+ httr = '1.4.7',
+ timechange = '0.2.0',
+ lubridate = '1.8.0',
+ isoband = '0.2.7',
+ tibble = '3.2.1',
+ ggplot2 = '3.3.6',
+ patchwork = '1.2.0',
+ later = '1.3.2',
+ promises = '1.3.0',
+ crosstalk = '1.2.0',
+ purrr = '1.0.2',
+ tidyselect = '1.2.1',
+ dplyr = '1.1.4',
+ tidyr = '1.2.1',
+ plotly = '4.10.4',
+ plyr = '1.8.7',
+ clipr = '0.8.0',
+ prettyunits = '1.1.1',
+ progress = '1.2.2',
+ tzdb = '0.4.0',
+ vroom = '1.6.3',
+ readr = '2.1.2',
+ reshape2 = '1.4.4',
+ plogr = '0.2.0',
+ RSQLite = '2.3.9',
+ systemfonts = '1.2.3',
+ textshaping = '0.3.6',
+ ragg = '1.2.5',
+ dbplyr = '2.2.1',
+ rstudioapi = '0.14',
+ dtplyr = '1.3.1',
+ backports = '1.4.1',
+ broom = '1.0.1',
+ processx = '3.8.5',
+ callr = '3.7.3',
+ reprex = '2.0.2',
+ modelr = '0.1.9',
+ conflicted = '1.2.0',
+ rematch2 = '2.1.2',
+ gargle = '1.3.0',
+ rematch = '1.0.1',
+ cellranger = '1.1.0',
+ ids = '1.0.1',
+ googledrive = '2.0.0',
+ googlesheets4 = '1.0.1',
+ readxl = '1.4.1',
+ selectr = '0.4-2',
+ xml2 = '1.3.3',
+ rvest = '1.0.3',
+ forcats = '0.5.2',
+ haven = '2.5.1',
+ tidyverse = '1.3.2',
+ doParallel = '1.0.17',
+ MatrixGenerics = '1.6.0',
+ svglite = '2.1.0',
+ svMisc = '1.2.3',
+ xts = '0.12.2',
+ TTR = '0.24.3'
+)
+for (pkg in names(special_versions)) {
+ tryCatch({
+ remotes::install_version(pkg, version = special_versions[[pkg]], repos = 'https://cran.r-project.org', quiet = TRUE)
+ }, error = function(e) {
+ message('Note: install_version for ', pkg, ' failed, trying install.packages')
+ install.packages(pkg, quiet = TRUE)
+ })
+}
+EOFSPEC
+
+RUN R --vanilla --slave --file=/tmp/install_special.R
+
+# Install development dependencies
+RUN cat > /tmp/install_dev_dependencies.R << 'EOFDEV'
+options(repos = c(CRAN = 'https://cran.r-project.org'))
+dev_dependencies <- c('cffr', 'covr', 'goodpractice', 'here', 'lintr', 'pkgdown', 'rcmdcheck')
+
+for (pkg in dev_dependencies) {
+ tryCatch({
+ remotes::install_version(pkg, repos = 'https://cran.r-project.org', quiet = TRUE)
+ }, error = function(e) {
+ message('Note: install_version failed for ', pkg, ', trying install.packages')
+ install.packages(pkg, quiet = TRUE)
+ })
+}
+EOFDEV
+
+RUN R --vanilla --slave --file=/tmp/install_dev_dependencies.R
+
+# Verify that all dependencies from DESCRIPTION are installed
+RUN cat > /tmp/check_description_deps.R << 'EOF'
+options(repos = c(CRAN = 'https://cran.r-project.org'))
+# Read the DESCRIPTION file
+desc_file <- "/opt/SCWorkflow/DESCRIPTION"
+if (!file.exists(desc_file)) {
+ stop("DESCRIPTION file not found at ", desc_file)
+}
+# Parse dependencies from DESCRIPTION
+desc <- read.dcf(desc_file)
+deps <- unique(c(
+ strsplit(desc[1, "Imports"], ",")[[1]],
+ strsplit(desc[1, "Suggests"], ",")[[1]],
+ strsplit(desc[1, "Depends"], ",")[[1]]
+))
+deps <- trimws(deps)
+# Check if each dependency is installed
+missing <- deps[!vapply(deps, requireNamespace, quietly = TRUE, FUN.VALUE = logical(1))]
+if (length(missing) > 0) {
+ stop("The following dependencies are missing: ", paste(missing, collapse = ", "))
+} else {
+ message("All dependencies are installed.")
+}
+EOF
+
+RUN R --vanilla --slave --file=/tmp/check_description_deps.R
+
+COPY Dockerfile /